As a lab we are interested in the following areas:
- Mechanisms of beneficial mutations
- Evolution during Infections
- Ecological-evolutionary dynamics
- Adaptive dynamics within host-associated microbiomes
- EvolvingSTEM
1. Mechanisms of beneficial mutations
For years, researchers like us have used microbes as model systems to understand how evolutionary processes work. Increasingly, we can also use evolution to understand how microbes work, as a potent genetic screen for mutations that increase fitness in any imaginable environment. These mutations affect genes and systems that often yield the best adaptations and usually teach us new, surprising, fundamental attributes of microbial biology. For example, we discovered that nonsynonymous mutations in the essential tRNA-modifying gene tilS that nearly eliminate its primary function somehow increase fitness in certain environments, pointing toward a new secondary function. We’ve also identified novel mechanisms of antimicrobial resistance and new regulatory functions of enzymes involved in cyclic-di-GMP turnover that enable functional diversity in biofilms. More recently we’ve collaborated with colleagues who have developed animal models for experimental evolution, which has led us to discover how certain beneficial mutations produce adaptations to the host environment. We continue to investigate why mutations in certain genes are repeatedly selected in evolution experiments or chronic infections and how they work.
This work is supported by grants from NSF and NIH.
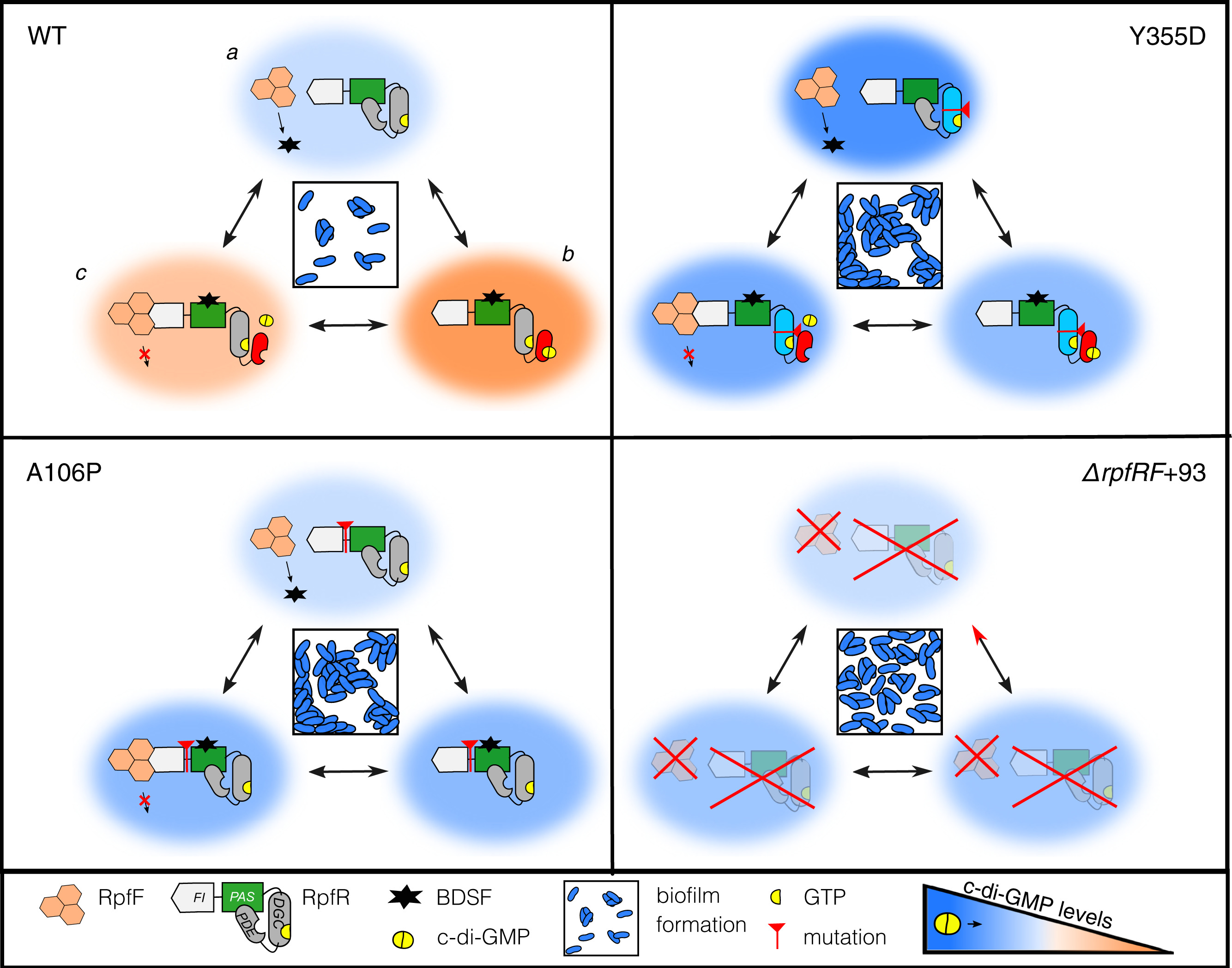
Possible ways rpfR mutants can influence biofilm phenotypes. Figure from Mhatre et al. PNAS, 2020
2. Evolution during Infections
Microbes often adaptively evolve when establishing infections by escaping components of the immune system and subsequently, antimicrobial treatment. We are interested in how bacterial populations evolve to escape antibiotic and host pressures during infections, with the goal of improving prediction of antimicrobial resistance (AMR). We study the evolution of drug resistance and pathogenicity in near real time through in vitro and in vivo experimental evolution using comparative genomics and analyses of longitudinal deep-sequenced populations. Our research questions include: (1) mechanisms of drug resistance and treatment failure, (2) the impact of the biofilm lifestyle on resistance, (3) how host immune state influences resistance evolution, and (4) comparative genomics of multi-drug resistant clinical isolates.
This work is supported by the NIH U19AI158076 CARBIRU project and by the PA Department of Health.
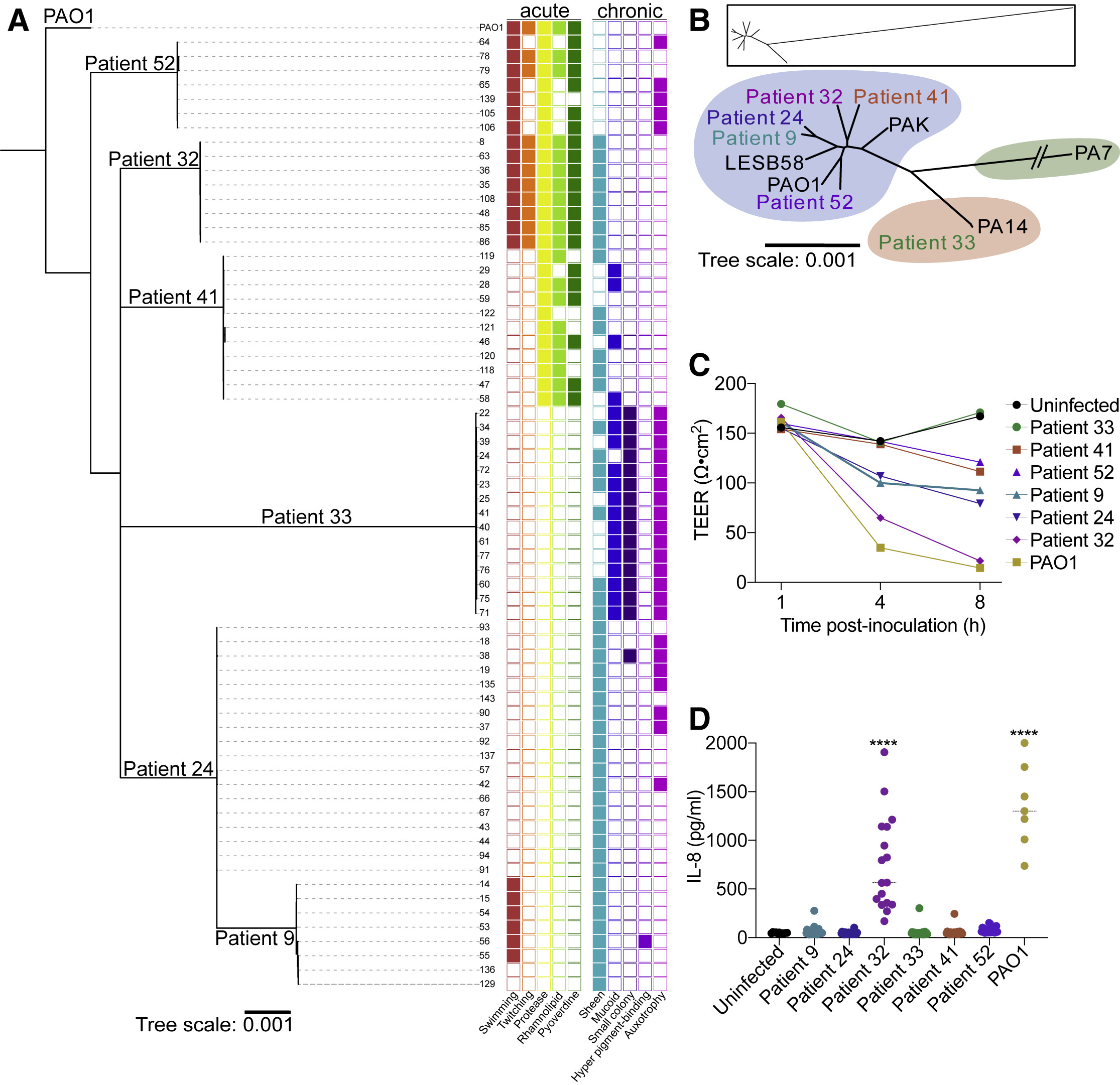
Pseudomonas aeruginosa from sinuses of people with cystic fibrosis show diverse pathoadaptive traits. Figure from Armbruster et al (2021) Cell Reports.
3. Ecological-evolutionary dynamics
Microbes exist within complex communities where both abiotic factors and inter-/intra-species interactions influence evolutionary trajectories. These environmental factors impose selective forces on communities. Likewise, individuals in a community can modify their environment and indirectly influence selective pressures applied to the community. We are interested in understanding such ecological and evolutionary dynamics at a community scale, especially in the context of biofilms, polymicrobial communities, and active bacteriophages.
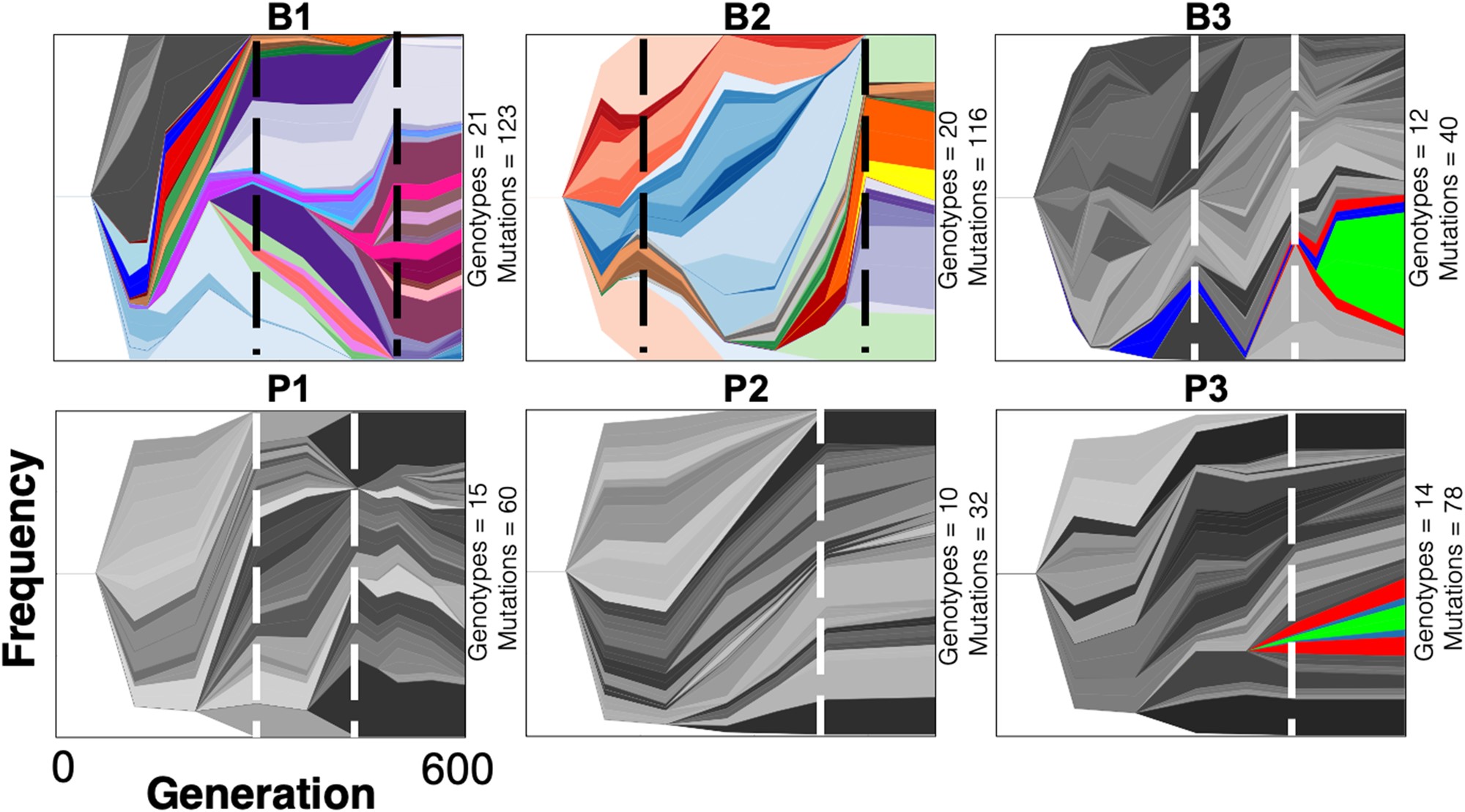
Muller plots showing the changes in Pseudomonas aeruginosa genotype frequency over time. Muller plots were created in Lolipop, a bioinformatic tool developed in our lab. Figure from Harris, Flynn, & Cooper (2021) Mol Biol Evol.
4. Adaptive dynamics within host-associated microbiomes
Like any disrupted ecosystem, when a host-associated microbiome is far from equilibrium, certain community members may be selected to inhabit vacant niches in which they are relatively maladapted. These lineages may undergo rapid adaptive evolution that can be tracked by longitudinal genome sequencing, alongside more typical unbiased measures of the community as it remodels. We study these processes in a wide range of systems with experts focused on particular diseases or symbioses, including:
- in the upper and lower airways of persons with cystic fibrosis,
- during establishment of chronic wounds,
- during chronic infections of prosthetic joint replacement
- during colonization of the infant gut
- during symbiosis with the Hawaiian bobtail squid and even among the virome within bacterial cells
(NIAID F31AI179118, NHLBI R33H137077 w/ Bomberger, R01DK120697 w/ Hand, R01AI165519 w/ Van Tyne, R01HL162964 w/ Zemke)
5. EvolvingSTEM
We created EvolvingSTEM, a research-education partnership program that uses evolution-in-action to capture the imagination of middle and high school biology students. We partner with classrooms across the US to engage students in an authentic laboratory experience. They evolve populations of Pseudomonas fluorescens under selection for biofilm growth and observe conspicuous changes to colony morphology in as little as one week. Our goals are to (1) increase understanding of key life science topics, including evolution, microbiology, and genetics, (2) provide training in essential biotechnology skills, and (3) inspire an enduring interest in science. Students gain confidence in STEM topics through hands-on experiences and drawing connections from their bench-top experiments to real-life medical applications. In addition, we train teachers to effectively implement our program in their classrooms through an immersive 8-week summer research experience hosted in our laboratory through support from the National Science Foundation (NSF BIORETS 2147075, NIH SEPA R25AI180989).

High school students doing hands-on evolution experiments in class through the EvolvingSTEM program.
Interested? Learn more at the EvolvingSTEM’s official website